May 2017
DR THABISO MOTAUNG, ARC-Small Grain, Bethlehem
Plant-pathogenic fungi cause diseases to major crop plants and threaten global food security. Virulence genetics underlie these fungal diseases and plant host colonisation. Therefore, genetic determinants are mostly to blame for the onset of pathogenesis, crop failures and subsequent staple food price hikes.
As such, pathogenicity studies can use collections of fungal strains containing mutant genes to provide immense function-based information regarding virulence genes of economically relevant fungi. These fungal mutant collections can assist a search for potential fungicidal targets and novel resistance genes through fungal effector search.
This article provides an updated assessment of the function-based contribution of mutant collections, using Magnaporthe oryzae and Fusarium graminearum as pathogenic fungal models.
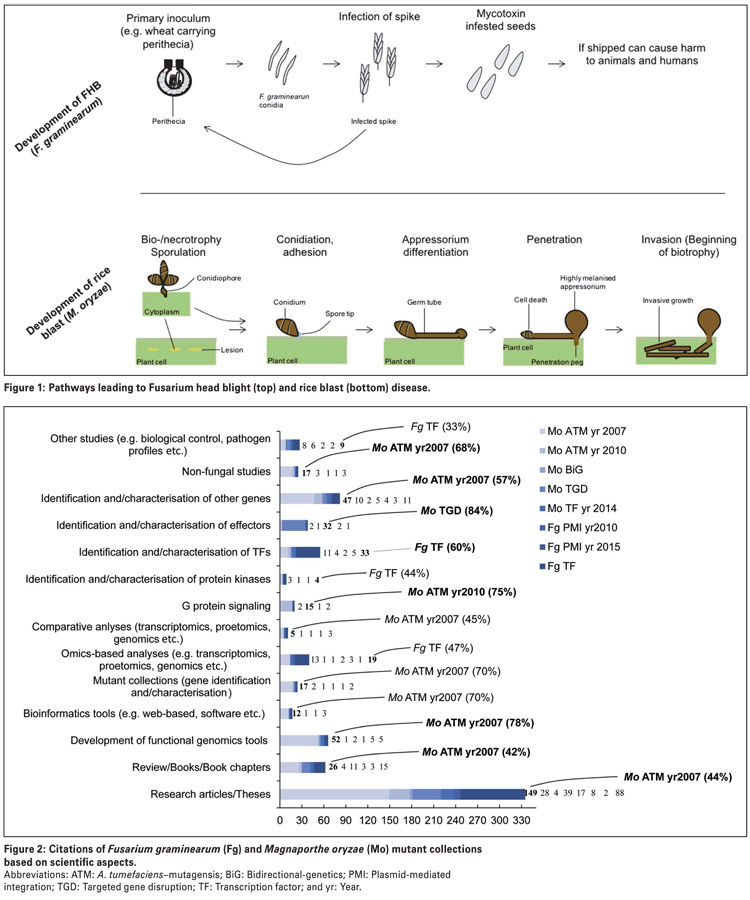
Fusarium graminearum is a predominant pathogen of head scab or Fusarium head blight in wheat and barley, and a cause of stalk and ear rot infections in maize (Photo 1). 
These crops are considered important South African crops, which makes F. graminearum a relevant pathogen to study in this country.
Fusarium graminearum also produces several important fungal toxins, including commonly studied toxins, such as deoxynivalenol, nivalenol and zearalenone. When released, these toxins are often problematic for many reasons in that they accumulate post-harvest and potentially contaminate food and feed, which in turn, present a health hazard to animals and humans.
Therefore, studying genes which are associated with the synthesis and release of these toxins will essentially lead to effective control strategies against Fusarium head blight epidemics and associated health impacts.
Fusarium graminearum mutant collections are therefore a promising molecular genetics tool for the high-throughput discovery of genes associated with mycotoxin production and regulation.
Magnaporthe oryzae is a leading fungal pathogen of rice in Asia. Although not largely known to infect crops in South Africa, M. oryzae has destroyed about 15 000 ha of wheat in Bangladesh, suggesting that the underlying disease, called wheat blast, is likely to emerge in wheat growing regions of South Africa in future.
This pathogen elaborates an asymptomatic infection phase, called biotrophy, that allows it to grow and feed on living plant tissues (Figure 1), and is also able to switch to the symptomatic phase called necrotrophy.
Additionally, M. oryzae can secrete proteins called effectors, which, in addition to interacting with plant resistant genes, can induce host cell death. However, host cell death induced by fungal effectors may be host-specific.
The ability of M. oryzae to switch from the biotrophic to the necrotrophic phase means it can feed on both living and dead plant tissues, which means this fungus is neither a biotroph nor a necrotroph, but instead is called a hemibiotroph.
Magnaporthe oryzae is one of the few hemibiotrophs that are experimentally tractable and has for this reason been subjected to molecular genetics tools, such as large-scale mutant generation.
The advantage of fungal collections is that the same collections can be screened for any phenotype of interest using a range of culture conditions and host plants. For the past decade, successful mutant screens have been conducted in various background strains of F. graminearum and M. oryzae (Table 1).
Such screens of mutant collections have produced vast amounts of data, much of which has advanced knowledge of parasitic mechanisms used by fungi to infect their host plants. As a result of these large-scale genetic studies, numerous genes have been assigned roles in the biological processes of fungal pathogens.
Owing to the importance of these genetic resources, a scientific literature study, focusing on papers looking at virulence gene discovery, modes of pathogenicity and regulation, and host-pathogen interactions was conducted. A more in-depth analysis involved splitting of papers citing mutant collections into twelve different scientific aspects (Figure 2).
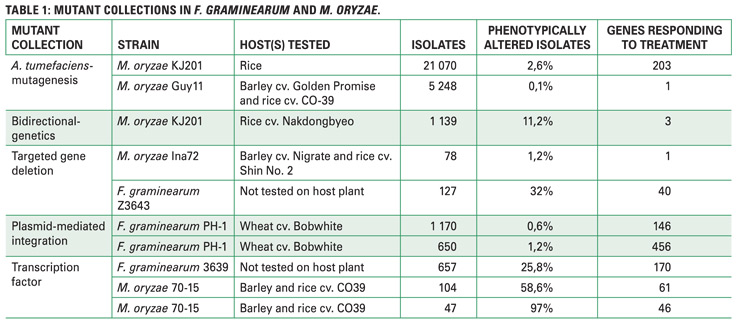
The results produced ten relevant mutant screens, a striking observation of which was that a large number of generated mutant isolates represented only a portion of phenotypically altered isolates; these phenotypically altered isolates are the very fungal strains containing mutations in specific genes, while the remainder of the genome is either uninterrupted or less important for fungal functions.
However, given that a few mutant isolates were altered, a few genes responding to different treatments were thus represented (Table 1). This may be due to dependence on less sensitive phenotypic assays and lack of additional phenotypic or in planta screens.
Citations of fungal mutant collections were assessed, the relevance of which is crucial to see if findings from these collections are channelled to solve agricultural problems in the South African context. Interestingly, none of the reports came from South Africa when a total of 397 citing papers, 116 citing F. graminearum mutant screens and 281 citing M. oryzae mutant screens (Figure 2), were screened.
Therefore, there is a huge gap to generate and screen fungal mutant collections in South Africa. Ultimately, performing such mutant screens, more in particular for F. graminearum, will drive the identification of virulence genes relevant to crops in this country. Furthermore, increasing the number of genes and related pathways involved in virulence will potentially complement the ongoing breeding efforts made for the South African producer.
To conclude, it is apparent that the application of fungal mutant collections is still in infancy when it comes to effector search. Only M. oryzae targeted gene disruption (TGD) collection provides screening of effectors. Available transcription factor (TF) mutant collections can potentially guide the development of alternative and effective fungicides, which target conserved TF proteins.
Since TFs control multiple genes which influence multiple biological processes, associated developmental costs of fungicides which target these TFs will substantially drop. This is because such fungicides will not be developed for countless downstream genes of TFs. Disease control procedures should thus unify findings from effector and TF mutant collections to advanced effector-assisted breeding and biological control using TFs as potential targets. The prospect of making collections of mutants for the South African molecular pathologist is within reach, and will potentially involve the inclusion to the list of mutant collections of fungi such as wheat rusts. 
Having these genetic resources in the country can greatly facilitate and accelerate disease combat efforts as well as significantly improve crop production and profitability for producers.
For more information, contact Dr Thabiso Motaung at motaungte@arc.agric.za.
Publication: May 2017
Section: On farm level

















